Shortest path algorithms have many applications in real life problems, such as:
- Finding the fastest route between two locations (Google Maps, GPS, etc.)
- In networking, finding a path between two nodes with the maximum possible bandwidth (represented by the minimum-weight edge along the path)
- Finding an optimal sequence of choices for reaching a certain goal state.
- Seam Carving
- VLSI design, etc.
Definitions
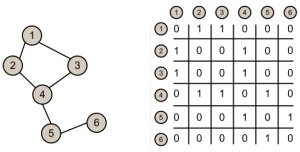
Let G = (V, E) represent a graph.
Let w : E -> W be a function that associates to each edge a cost (weight). If an edge does not exist between two nodes we assume a cost of +infinite.
Let numNodes represent the number of vertices in the graph.
Let numEdges represent the number of edges in the graph.
Let minDist[u] refer to an algorithm’s estimation of the minimum distance from the source to node u.
Let parent[u] refer to the second-to-last element on the minimum cost path from the source to node v.
The general problem asks us to find a path between two given nodes such that total sum of the weights is minimum.
Relaxing an edge
Relaxation of an edge (u, v) refers to improving the current estimation of the minimum cost for a path from the source node to node v by using the respective edge.
void relaxEdge (Vertex u, Vertex v)
{
if (minDist[v] > minDist[u] + w(u, v)) {
minDist[v] = minDist[u] + w(u, v);
parent[v] = u;
}
}
Single source shortest path problem
Given one source node, find the shortest path to all the other nodes in the graph. Ideally, we want to analyze each node/edge only once.
Scenarios:
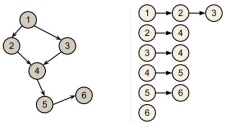
- The graph is undirected and acyclic (a tree)
In this situation there is only one path from one node to another so we can find the answer with a depth first search.
Time complexity: Θ (numNodes + numEdges)
- The graph is a directed and acyclic (a DAG)
For solving the problem we can explore each node in sorted topological order and apply the relaxEdge procedure for each corresponding edge. This approach requires only two traversals of the graph.
Time complexity: Θ (numNodes + numEdges) - A graph where each edge has an equal weight.
For this scenario, the best approach is to apply the Breadth First Search algorithm. The order in which nodes are explored is given by the minimum number of edges from the source, which coincides with the shortest path.
Time complexity: Θ (numNodes + numEdges)- A graph with edges that have only positive weights.
When we extracted a node from the front of the queue during BFS, we had the guarantee that it was the closest unexplored node from the source, in terms of number of edges. If we change the problem so that edges have different but positive weights we need to replace the queue with another data structure that supports efficient interrogation for the closest node. This type of data structure is called a priority queue. We would also like to be able adjust the estimated value for a node efficiently, for when we relax an edge.
The following algorithm is known as Dijkstra’s algorithm.
// setup an initial distance estimation void initDistance(Graph, Vertex s) { for each Vertex v in Graph { minDist[v] = infinite; visited[v] = false; } // set minDist for source to 0 minDist[s] = 0; } void Dijkstra(Graph, Vertex source) { initDistance(Graph, source); PriorityQueue PQ; PQ.add(source); while (!PQ.empty()) { // u is the closest unexplored node to the source Vertex u = PQ.extractMin(); mark u as explored for each Vertex v that is connected to Vertex u { if Vertex v is not explored { relaxEdge(v, u) updatePQ(v); // update PQ with the new estimation // of distance for v } } } }For Java and C there already exist implementations for a priority queue data structure: PriorityQueue and priority_queue. These implementations have logarithmic time complexity for element addition and extraction, but don’t allow you to update the priority of a random element.
Note that, since the priority queue will contain node references, to use these existing data structures you need to implement a comparator first and supply it to the constructor.
The default Priority Queue in Java works as a min-heap (returns the minimum element, as indicated by the comparator), while the C++ equivalent data structure priority_queue is a max-heap (default behaviour is to return the maximum element).
class NodeComparator implements Comparator { @Override public int compare(Vertex u, Vertex v) { return minDist[u.index] < minDist[v.index] ? 1 : -1; } }Complexity of Dijkstra’s algorithm
We explore each node exactly once and this translates to one call to PQ.extractMin() for each node. We also only check each edge once, but in the worst case scenario we have to update the priority queue structure with each check.
Therefore, we can express the complexity as:
Θ (numNodes * Extract-Min + numEdges * UpdatePQ)
The exact complexity of Dikstra’s algorithm depends on how we implement the priority queue:
As a simple array:
- Extract-Min: Θ (numNodes)
- UpdatePQ: Θ (1)
Time complexity: Θ (numNodes^2 + numEdges), which is efficient for dense graphs.
As a binary heap:
- Extract-Min: Ο( log(numNodes) )
- UpdatePQ: Ο( log(numNodes) )
Time complexity: Θ (numEdges * log(numEdges)), which is efficient for sparse graphs.
As a Fibonacci heap:
- Extract-Min: Ο( log(numNodes) )
- UpdatePQ: Ο(1)*
Time complexity: Θ (numNodes * log(numNodes) + numEdges)
Possible exam questions
- What is the order of exploration of the nodes, using Dijkstra’s algorithm, given a graph and a source node?
- Why doesn’t Dijkstra work if we have negative weight edges? Think of a simple example.
- A graph with edges that have only positive weights.
- A graph with edges that have both negative and positive weights
For this scenario we may apply the Bellman Ford algorithm, where we basically relax each edge for numNodes-1 iterations:void BellmanFord(Graph, source) { initDistance(Graph, source); for (i = 1; i <= numNodes - 1; i++) { for each edge(u, v) in Graph { relax(u, v); } } }Complexity for Bellman Ford: Θ (numNodes * numEdges).
Possible exam question:
If it is still possible to relax an edge after we run Bellman-Ford than there exists a negative cycle in the graph.Why? Hint: How many iterations does it take to obtain the shortest distance for any destination in the worst case scenario?

 Posted by raduiacob
Posted by raduiacob